Archive \ Volume.11 2020 Issue 3
Antimicrobial Resistance, Biofilm Formation and Virulence Factors in Enterococcus faecalis Strains Isolated from Urinary Tract Infection in Kermanshah, Iran
Mahmoud Shahveh 1, Elahe Tajbakhsh 1*, Hassan Momtaz 1, Reza Ranjbar 1, 2
1 Department of Microbiology, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran. 2 Professor of Medical Bacteriology, Molecular Biology Research Center, Systems Biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran
Abstract
Aims and Objectives: This study aims to specify the antimicrobial resistance pattern and virulence genes of Enterococcus faecalis isolated from individuals suffering from an infected urinary tract in Kermanshah, Iran. Patients and methods: Isolates were gathered from 1000 urine samples of patients diagnosed with the infected urinary tract. Biofilm assays were performed by microtiter plate test through reading the OD490 following the crystal violet staining. The Kirby–Bauer disc diffusion method was performed for the antimicrobial susceptibility testing. PCR reaction was applied to study the virulence factors. Results: Of 1000 urine samples, E. faecalis was reported 5%. Strong, moderate and weak biofilm reactions reported 80%, 12%, and 8% respectively. The most resistance reported to cotrimoxazole, vancomycin and amikacin and no resistance to nitrofurantoin were reported. Prevalence of efe A, ace, gel E, esp, cyl M, agg, cyl A and cyl B in strong biofilm formation isolates was reported 100%, 87.5%, 82.5%, 80%, 60%, 57.5% 37.5% and 30% respectively. There was a significant relationship between the frequency of efa A, ace, gel E, esp genes and strong biofilm reaction (p<0.0001). Conclusion: The presence of E. faecalis strains resistant to co-trimoxazole and vancomycin with some virulence factors is alarming the researchers. Since antibiotic resistance genes are probably transmitted among enterococci, and Staphylococci, controlling infections made by enterococci as well as the appropriate administration of antibiotics could treat the nosocomial infections effectively.
Keywords: Antibiotic Resistance; Enterococcus faecalis; Urinary Tract Infection; Virulence genes
 INTRODUCTION
INTRODUCTION
Enterococci as Gram-positive and catalase-negative cocci can develop along with 6.5% salts and 40% bile salts [1]. The most common enterococci found in human infection are Enterococcus faecalis (85-90%) and Enterococcus faecium (5-10%). These bacteria belong to the normal flora of the human digestive system having been used commensal bacteria in the past. Enterococci is capable of triggering infections in wounds such as burn wounds, urethra, and even endocarditis [1]. Investigations show that biofilm is produced by E. faecalis and biofilm growth is under the control of its quorum-sensing system [1]. Microbial biofilm is a bacterial mass primarily related to biotic and abiotic surfaces that trigger the cells to bind irreversibly to the surfaces by making extracellular polymers and building a matrix of alginate [2]. It is believed that there is an association between the development of biofilm with bacterial resistance to antibiotic treatments that probably would lead to acute problems in this area. Bacteria taking part in biofilms behave differently compared to the planktonic cells [3, 4]. The extracellular surface protein (esp) increases the ability to colonize and create bacterial biofilm in vitro, and it seems to be related to the presence of biofilms in vivo [5]. Ebp protein (endocarditis and biofilm-associated pili) is one of the most important proteins encoded in the pathogenesis of E. faecalis. In the Ebp operon, the Ebp A protein plays a major role in the pathogenesis process. Also, it has been verified that Ebp protein is important for the formation of biofilms so that it can cause urinary tract infection and endocarditis in the experimental models [6]. Some enterococcal virulence factors have been recognized such as adhesions and secreted virulence factors. The main adhesion factors are Asa (aggregation substance), Esp (extracellular surface protein), EfaA (E. faecalis antigen A), Ace (adhesin of collagen from E. faecalis) and Ebp [6]. Proteins like hemolysin, gelatinase, and aggregation substances are also involved in the plasmid exchange systems. The severity of endocarditis and endophthalmitis in the animal model is aggravated significantly following the production of cytolysin leading to severe enterococcal diseases in humans [7]. The initial bonding and the formation of the E. faecalis biofilm are accompanied by the presence of the enterococcal surface protein. The chromosomal gel E gene encodes the gelatinase. This enzyme is an extracellular metalloproteinase that hydrolyzes collagen, gelatin, and small peptides; it also is involved significantly in the formation of animal endocarditis. The role of cytolysin in the onset of the disease has been verified by Epidemiological investigations [8]. Resistance to a wide range of antibiotics is observed in enterococci, such as inherent resistance to macrolides, chloramphenicol, penicillin, tetracycline, and aminoglycosides. Multiple drug resistances are among the characteristics of enterococci [3]. Resistance genes may be disseminated in bacteria through the interference of the horizontal gene transfer. Also, selective pressure by drugs has significant involvement in resistance dissemination; besides, enterococci are also important for wide-range antibiotic resistance. In this respect, genetic mobile elements like conjugative plasmids and transposons can spread resistance genes [4]. This study aims to specify the antimicrobial resistance pattern and virulence genes of Enterococcus faecalis isolated in UTI patients in Kermanshah, Iran.
METHODS
Ethics and consent
In the current study, we tried to protect the life, health, dignity, integrity, rights to self-determination, privacy, and confidentiality of personal information of research subjects. We conform to generally accepted scientific principles based on a thorough knowledge of the scientific literature, other relevant sources of information, and adequate laboratory and, as appropriate, animal experimentation. All samples were taken from volunteer patients for this research. All ethical issues were considered and this research was performed with hospitals’ permission. The name and identity, personal information and even patients’ illnesses and their medical information remained secret. All of the patients voice their satisfaction to use their sample in this investigation, especially to determine antibiotic resistance in UPEC strains.
This thesis has been approved in Islamic Azad University Shahrekord branch with code 13330507962017.
Sampling, Isolation, and Identification
In this cross-sectional study, approximately 1000 urine samples were gathered from successive outpatients with a UTI in Kermanshah, from May to August 2017. To separate the bacteria, an early morning midstream urine samples were cultured on blood agar containing 5% sheep blood under sterile conditions and incubated for 24 hr at 37°C. To verify the suspected colonies of enterococci, E. faecalis ATCC 29212 (Pasteur Institute of Iran) was applied as a standard strain. The pure cultures of suspected colonies were sub-cultured on Bile Esculin agar and incubated for 48 hr. at 37°C. Moreover, Gram stain, catalase test, growth at 6.5% NaCl and PYR test were performed for the early identification of enterococci. In this study, the arabinose fermentation test was employed to differentiate E. faecalis and E. faecium [9, 10].
Microtiter Plate Assay for detection of biofilm
The modified crystal violet staining method was applied to quantify the formation of biofilm on the polystyrene microplates. For each strain, few colonies were suspended in physiological saline to 0.5 McFarland and vortexes for 1 min. 180 µL Trypticase soy broth (TSB) + 0.5% glucose and 20 µL of bacteria suspension was used to fill 96 well polystyrene Microtiter plates. All plates were performed in duplicate. Negative controls were TSB +0.5% glucose alone. Following the stationary aerobic incubation for 24 hr. at 37 0C and 5% CO2, the broth was cautiously taken away and 300 µL of sterile phosphate-buffered saline (PBS, room temperature) was used to wash the well threefold. 150 µL methanol for 20 min was used to fix the biofilm; then, they were put to dry in the air in an inverted position in the warm room (around 30 min). To stain the biofilms, 150 µL of crystal violet solution in water (2%) was used for 15 min at room temperature; then, the wells were put under the running water tap for rinsing. To invert the Microtiter plates, they were put on a paper towel and air-dried. To specify the yield of biofilms, 150 µL of 33% acetic acid was added to each well to destain the biofilms and plates covered with lids were placed at room temperature for half an hour with no shaking. The optical density (OD) was measured at 490 nm in a spectrophotometer. This way, the isolates with OD equal to or higher than 0.216 were taken as strong biofilms, if they had an OD between 0.54 and 0.108, as weak biofilms, if they had an OD less than 0.54 as negative biofilms [11]. DNA of the samples that were taken as positive and negative biofilms was extracted using a DNA extraction kit.
Antimicrobial Susceptibility Testing
The Kirby–Bauer disc diffusion method was applied using Mueller–Hinton agar (Merck, Germany) according to the Clinical Laboratory Standards Institute (CLSI) guidelines to conduct the antimicrobial susceptibility testing [12]. Some sorts of antibiotics with proper disks containing vancomycin (VAN 10µg), co-trimoxazole (SXT 25µg), ceftazidime (CAZ 30μg), norfloxacin (NOR 10μg), amikacin (AN 30μg), gentamicin (GM 120μg), cefotaxime (CTX 30 μg) and nitrofurantoin (FM 300μg) (produced by PadTan-Teb, Iran).
DNA extraction and PCR Assay
A DNA extraction kit (Cinapure DNA, CinaClon, Iran) was applied according to the manufacturer’s instructions to extract genomic DNAs from E. faecalis isolates. Based on the method specified by Sambrook and Russell [13], the total DNA was measured at 260 nm optical density. PCR was applied using specifically targeted primers to detect enterococci [12].
Identifying the virulence genes by PCR method
PCR was applied using specific primes to identify the virulence factors genes including gel E, esp, agg, ace, cytolysin (cylM, cylA and cyl B) genes and ebp A, ebp B, ebp C. Tables 1 and 2 show the applied primer sequence, the annealing temperature, and the used PCR program. A DNA thermal cycler was used to perform PCR, (Master Cycler Gradient, Eppendorf, Germany). Ethidium bromide was used to stain the amplicons that were then electrophoresed in 1.5% agarose gel at 80 V for 30 min. To observe and photograph PCR products, the UV doc gel documentation systems (Uvitec, UK) was used. A comparison was performed between the PCR products against a 100 bp DNA marker (Fermentas, Germany) [14-17].
|
Table 1: The oligonucleotide primers and the Multiplex PCR programs used for amplification of virulence genes of E. faecalis isolates |
||||
|
Target gene |
Primer Oligonucleotide sequences (5’-3’) |
Accession Number |
Size of amplicon (bp) |
Annealing Temperature (oC) |
|
tfu |
F: TCAAGTACAGTTAGTCTTTATTAG R:ACGATTCAAAGCTAACTGAATCAGT |
CP015883.1 |
112 |
59 |
|
Ebp A |
F: CTAACAAAAATGATTCGGCTCCAG R: ATCTCACGCATTTTATCTTCAACT |
CP028285 |
517 |
60 |
|
Ebp B |
F: CTGAAGGAAAAACGGTCCAA R: CTTTTGCGTCGTCAGTGTGT |
CP022059 |
1003 |
55 |
|
Ebp C |
F: GATAAATATCAAGGACTGGCAGA R: AAGCATACTCTCCAGAAGTCACG |
CP022059 |
600 |
58 |
|
asa |
F: CCAGCCAACTATGGCGGAATC R: CCTGTCGCAAGATCGACTGTA |
CP22712 |
419 |
51 |
|
Gel E |
F: ACCCCGTATCATTGGTTT R:: ACGCATTGCTTTTCCATC |
KU311665 |
629 |
51 |
|
Cyl A |
F: TGGATGATAGTGATAGGAAGT R: TCTACAGTAAATCTTTCGTCA |
CP015883 |
517 |
55 |
|
Cyl M |
F: CTGATGGAAAGAAGATAGTAT R: TGAGTTGGTCTGATTACATTT |
AY032999 |
742 |
55 |
|
Afa A |
F: GACAGACCCTCACGAATA R: AGTTCATCATGCTGTAGTA |
KY070337 |
705 |
55 |
|
Cyl B |
F: ATTCCTACCTATGTTCTGTTA R: AATAAACTCTTCTTTTCCAAC |
KU311664 |
843 |
54 |
|
ace |
F: GAGCAAAAGTTCAATCGTTGAC R:GTCTGTCTTTTCACTTGTTTCT |
AF159247 |
1003 |
54 |
|
agg |
F: AAGAAAAAGAAGTAGACCAAC R: AAACGGCAAGACAAGTAAATA |
CP002493 |
1553 |
54 |
|
esp |
F: TTGCTAATGCTAGTCCACGACC R: GCGTCAACACTTGCATTGCCGAA |
AF034779 |
933 |
54 |
|
Table 2: The Multiplex PCR programs used for amplification of E. faecalis isolates |
||
|
Gene |
PCR programe |
M-PCR Volume (50 μL) |
|
tfu |
1 cycle: 95 0C ------------ 5 min. 31 cycle: 95 0C ------------ 45 s 59 0C ------------ 60 s 72 0C ------------ 60 s 1 cycle: 72 0C ------------ 5 min |
5 μL PCR buffer 10X 2 mM Mgcl2 200 μM dNTP (Fermentas) 0.4 μM of each primers F & R 1 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
|
Ebp A |
1 cycle: 94 0C ------------ 5 min. 32 cycle: 94 0C ------------ 60 s 60 0C ------------ 60 s 72 0C ------------ 2 min 1 cycle: 72 0C ------------ 5 min |
5 μL PCR buffer 10X 2 mM Mgcl2 200 μM dNTP (Fermentas) 0.4 μM of each primers F & R 1 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
|
Ebp B |
1 cycle: 94 0C ------------ 5 min. 32 cycle: 94 0C ------------ 60 s 55 0C ------------ 60 s 72 0C ------------ 2 min 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 2 mM Mgcl2 200 μM dNTP (Fermentas) 0.4 μM of each primers F & R 1 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
|
Ebp C |
1 cycle: 94 0C ------------ 5 min. 32 cycle: 94 0C ------------ 60 s 58 0C ------------ 60 s 72 0C ------------ 2 min 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 2 mM Mgcl2 200 μM dNTP (Fermentas) 0.4 μM of each primers F & R 1 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
|
Asa, gel E |
1 cycle: 95 0C ------------ 5 min. 30 cycle: 95 0C ------------ 30 s 51 0C ------------ 30 s 72 0C ------------ 60 s 1 cycle: 72 0C ------------ 6 min |
5 μl PCR buffer 10X 2.5 mm Mgcl2 200 μM dNTP (Fermentas) 0.5 μm of each primers F & R 2 U Taq DNA polymerase (Fermentas) 3 μl DNA template |
|
Cyl A, cyl M, afa A |
1 cycle: 95 0C ------------ 6 min. 30 cycle: 94 0C ------------ 60 s 55 0C ------------ 60 s 72 0C ------------ 45 s 1 cycle: 72 0C ------------ 7 min |
5 μL PCR buffer 10X 2.5 mM Mgcl2 300 μM dNTP (Fermentas) 0.4 μM of each primers F & R 2 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
|
Cyl B, ace, agg, esp |
1 cycle: 94 0C ------------ 6 min. 35 cycle: 95 0C ------------ 60 s 54 0C ------------ 90 s 73 0C ------------ 45 s 1 cycle: 72 0C ------------ 7 min |
5 μL PCR buffer 10X 2.5 mM Mgcl2 300 μM dNTP (Fermentas) 0.4 μM of each primers F & R 2 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
Statistical analysis
SPSS software (Version 25.0) (IBM SPSS Statistics) was used for statistical analysis. Fischer exact was performed for data analysis. P-value < 0.05 was considered significant.
RESULTS
Out of 1000 Urine samples, E. faecalis was detected in 50 samples (5%). All samples tested positive microbiologically were tested positive in a molecular study conducted using a specific primer (Figure 1).
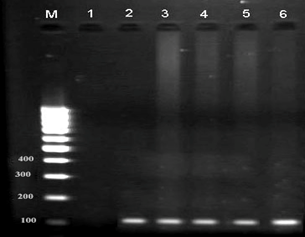
Figure 1. Result of the PCR Assay for identification of tfu gene in E. faecalis isolates. M: DNA size ladder 100 bp (Fermentas), lane 1: negative control; lane 2-6: positive samples
In the microtiter plate method, the formation of biofilm was observed in all isolates categorized as strong, moderate and weak and based on OD. In this study, the strong biofilm reaction was observed in 40 isolates (80%), moderate biofilm in 6 isolates (12%) and weak biofilm reactions were reported in 4 isolates (8%). Among the isolates producing strong biofilm, ebp A, ebp B, and ebp C genes were detected in 40 isolates (100%). Among the isolates producing moderate and weak biofilm ebp A, ebp B and ebp C genes were identified in 50% of isolates (Figure 2).
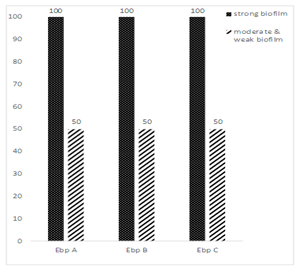
Figure 2: Prevalence of ebp A, ebp B and ebp C genes in biofilm-producing E. faecalis isolates
Figure 3 shows the resistance to the target antibiotics. The maximum resistance was against cotrimoxazole (100%), vancomycin (97.5%) and amikacin (72.5%) in strains producing a strong biofilm. Nevertheless, in moderate and weak biofilm formation strains, the maximum resistance was against cotrimoxazole (100%), vancomycin (90%), and amikacin (60%). In this study, resistance to nitrofurantoin was not recorded. According to the statistical analysis with Fisher exact, there antibiotic resistance and type of biofilm (p>0.05) were not associated significantly.
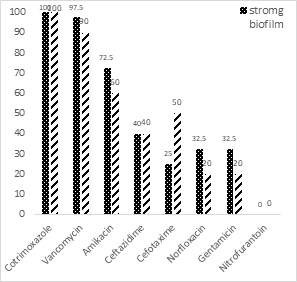
Figure 3: Antibiotic resistance pattern of the biofilm-producing E. faecalis isolates
Figure 4 shows how virulence genes are distributed among the clinical E. faecalis isolates. The most prevalence of virulence genes was in strong biofilm formation E. faecalis strains: efa A (100%), ace (87.5%), gel E (82.5%) and esp (80%). But in moderate and weak biofilm formation E. faecalis strains: efa A (20%), ace (20%), gel E (20%) and esp (10%). There was a significant relationship between the frequency of virulence genes of efa A, ace, gel E, esp and strong biofilm reaction (P<0.05). However, the frequency of virulence genes and moderate and weak biofilm formation were not associated significantly.
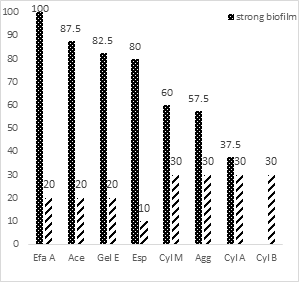
Figure 4: Prevalence of virulence genes in E. faecalis isolates
Result of the PCR assay for the identification of virulence genes that show in figures 5 to 8.
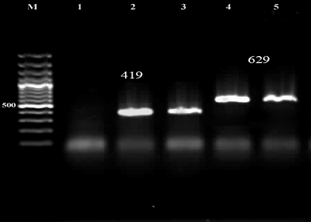
Figure 5. Result of the PCR Assay for identification of asa and gel E genes in E. faecalis isolates. M: DNA size ladder 100 bp (Fermentas), lane 1: negative control; lane 2,3: asa gene, lane 3,4: gel E gene
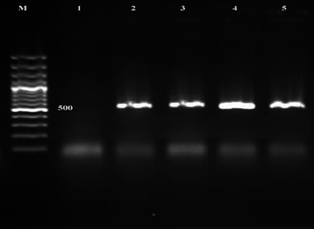
Figure 6. Result of the PCR Assay for identification of the ebp C gene in E. faecalis isolates. M: DNA size ladder 100 bp (Fermentas), lane 1: negative control; lane 2-5: ebp C gene
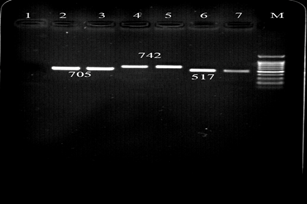
Figure 7. Result of the PCR Assay for identification of cyl A, cyl M and afa A genes in E. faecalis isolates. M: DNA size ladder 100 bp (Fermentas), lane 1: negative control; lane 2,3: afa A, lane 4,5: cyl M and lane 6,7 cyl A.
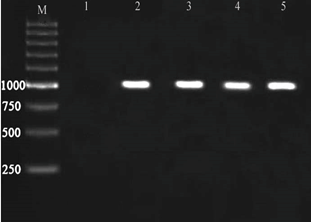
Figure 8. Result of the PCR Assay for identification of ebp B gene in E. faecalis isolates. M: DNA size ladder 100 bp (Fermentas), lane 1: negative control; lane 2-5: ebp B.
DISCUSSION
Enterococci are naturally resistant to several antibiotics; they are also capable of developing drug resistance either by chromosomal mutations, by transfer of plasmids, or transposon acquisition containing genetic sequences leading to resistance [18]. E. faecalis has come to be a common UTI pathogen. E. faecium outperforms E. faecalis in terms of drug resistance, but E. faecalis biofilm formation is more prevalent than that for E. faecium. The increased importance of enterococci as nosocomial pathogens can be to some extent justified by their intrinsic capability to acquire, accumulate, and share extrachromosomal elements encoding virulence traits or antibiotic resistance genes [19].
Our study showed a higher percentage of resistance to cotrimoxazole, vancomycin, and amikacin: 100%, 97.5%, and 72.5% in isolates that produced strong biofilm and 100%, 90%, 60% in isolates that produce moderate and weak biofilm. The high prevalence resistance to rifampicin and amikacin (86.2%) and erythromycin (73.9%) among enterococcal strains isolated from urinary tract infections have been reported by Sharifi et al. (2013) [20]. In the present study, resistance to amikacin was consistent with the results reported by Sharifi et al. In our study, resistance to nitrofurantoin was not observed which was probably due to one of the treatment options. However, Shahrakie et al. (2017) [4] reported that in the clinical multidrug resistance enterococci isolates in Southeastern Iran, resistance to nitrofurantoin was reported 7.93%. In the study by Kuhn et al. (2005) [3] resistance to vancomycin was reported 8-11%, but, our research resistance to vancomycin reported 98%. Their findings suggested significantly increased vancomycin resistance among clinical E. faecalis isolates from 2005 onwards. In our study, the biofilm formation of E. faecalis strains isolated from UTIs was explored in Kermanshah, Iran. Enterococci, (especially E. faecalis) are involved significantly in the production of biofilm in the development of UTI. A suitable milieu is brought about by biofilms for microbial survival within the host as the organisms are protected from the host immune response, as well as antibiotics and antimicrobial agents. Several studies have focused on key virulence genes of enterococci associated with biofilm formation in these bacteria2. In the present study, the strong biofilm formation was reported as 80%, moderate biofilms as 12%, and weak biofilm as 8%. Prevalence of ebp A, ebp B and ebp C genes in isolates producing strong biofilm was reported 100% and in isolates producing moderate and weak biofilm were 50%. Statistical analysis with Fisher exact test showed a significant correlation between biofilm formation and ebp genes (p<0.05). In a study by Talebi et al. (2015) [21] on 58 isolates of E. faecalis obtained from environmental sources and 32 isolates of Enterococcus faecalis obtained from two hospitals in Tehran, the ability to biofilm formation was investigated by microtiter plate. The capability of the isolates to join strongly was 62% and 71% for patient and environmental samples, in the respective order. In the hospital samples, ebp A was detected in 50 isolates (86%) and ebp B in 56 isolates (95%), and ebp C in 56 isolates (97%). However, in the environmental samples, ebp A was reported in 29 isolates (91%), ebp B in 30 isolates (94%) and ebp C in 29 isolates (91%). which is lower than our reports and previous reports. This discrepancy might be due to the presence of variable genetic make-up amongst the isolates within the ebp gene [22, 23]. Studies have shown that in most cases, the bacterial isolation and stability directly depend on biofilm yield in the urinary tract [24, 25]. More studies have revealed the pathogenesis factors of E. faecalis, each of which has a particular role in the development of the disease. These factors act concurrently leading to increased virulence and causing tissue degradation and invasion. The aggregation substance in the enterococci produced in response to sex pheromones that cause them to bind these bacteria together and to create a cell mass. It is also involved significantly in stimulating adhesion, cellular invasion, and degradation of myocardial and lung tissues [26, 27]. The enterococcal superficial protein has roughly the same functions as an aggregation substance. The agent of binding to collagen is involved significantly in the binding of bacteria to collagen and cellular laminin, and its mutation leads to decreased endocarditis and UTI formation. There is a correlation between the enterococcal surface protein encoded by the chromosomal esp gene increased pathogenesis, colonization, persistence in the urethra and biofilm formation. It has been shown that surface proteins are involved in colonization and survival of E. faecalis in UTI in animal models.
Esp has been observed in large amounts in the isolates of endocarditis and bacteremia, but it is rarely seen in fecal isolates of healthy people [5]. Some previous studies focused on the association between the virulence genes and biofilm formation and concluded that the presence of esp and gel E. esp help the colonization and persistence of infection within the urinary tract [28]. In the present paper, the highest frequency of virulence genes in strong biofilm formation isolates was observed in efe A (100%), ace (87.5%), gel E (82.5%) and esp (80%). However, in moderate and weak biofilm-producing isolates efe A, ace, gel E and esp were much less abundant. It was shown that strong biofilm formation and efe A, ace, gel E, and esp genes are correlated significantly indicating the importance of these genes in biofilm formation in E. faecalis isolates. Samadi Kafil et al.[27] investigated 196 isolates of Enterococcus and concluded that the most prevalent genes were efe A (93.36%), cyl A and ace (81.63%). Esp and gel E were much less frequent compared to our research. In the present study, the presence of esp was not correlated with biofilm-forming ability among Enterococcus isolates. A significant statistical relationship was found in the study of Samadi Kafil et al. between biofilm production and asa and efa A gene, which is similar to the results of our research. Meanwhile, it was shown that the production of biofilms and the esp gene are not correlated. Some studies recommended that the esp gene is not essential nor adequate for the production of biofilm in enterococci [29, 30]. Results on the role of the esp gene in biofilm formation are conflicting. Some authors believed biofilm formation is promoted by esp promotes biofilm formation; but, additional factors may contribute to biofilm formation in enterococci [31, 32]. Medeiros et al. reported that there are significant correlations between strong biofilm formation and ace and gelE genes in clinical strains [17]. Heikens et al. reported that Esp protein is not involved in biofilm production and bacterial colonization. Since cytolysin operon is closely associated with the esp gene, cytolysin does not have any role in biofilm production. However, some researchers believe that the esp gene plays a role in biofilm production [33]. Kristich et al. (2004) [34] reported that strains without enterococci surface protein contributed to the biofilm formation. Biofilm formation is independent of the Esp. Gelatinase increases the formation of biofilm in E. faecalis. It has also been shown that Esp was not necessary for biofilm formation, but its existence has been necessary for the formation of large amounts of biofilm [35, 36]. Seno et al. (2005) [37] reported that the presence of Esp, gelatinase, and the ability of strains to biofilm formation are correlated in the in vitro conditions. In a study by Gozlan et al. on 55 isolates of E. faesium, 41 (75%) were positive and 14 (25%) were negative for the virulence genes tested. The esp gene was observed in 100% of all isolates taken from urine samples [28]. In our study, esp was present in 66% of all isolates. Virulence factors such as hemolysin and cytolysin exist in 32% of enterococcal species. It has been shown that the severity of endocarditis and endophthalmitis in the animal model is aggravated considerably by Cytolysin production leading to increased incidence of enterococci in humans [3]. The chromosomal gel E gene encodes the gelatinase enzyme that is an extracellular metalloprotease hydrolyzing collagen, gelatin, and small peptides and is involved significantly in the endocarditis formation in animal models. Gelatinase damages the host tissue and decreases the immune response, and contributes to the activation of the autolysins and degrading the peptidoglycans, and subsequently releasing DNA and forming biofilms. E. faecalis with gelatinase genes have been identified in about 33% of patients with endocarditis. Sharifi et al, (2013) [20], Heidari et al. (2016) [30] and Sabia et al. (2008) [31] reported gel E as the most recurrent virulence factor in E. faecalis strains, whereas some studies have shown the absence or low rate of gel E gene in enterococcal isolates [38-40]. In a study by Zhengv et al. (2018) on 113 isolates of E. faecalis, biofilm production was reported in 50.40%, which is lower than other studies. In this study, it was shown that gel E is effective in biofilm production. However, biofilm production and esp gene [41] were not correlated. Nonetheless, others also reported that gel E is not involved in the production of biofilms. In the study by Zhengv et al. the production of biofilm in gel E negative isolates more than in positive gel E isolates. In our study, there was an association between the production of biofilm and frequency of gel E. The cyl A and ace genes were not detected by Gozlan et al. and this is not consistent with our findings [28]. In our study, the ace gene was reported in 74%, cyl M 54%, cyl A 50%, and cyl B 22% and there was a relationship between the production of biofilm and frequency of ace gene. The most frequent virulence genes reported by Shokohizadeh et al. (2018) among 56 enterococci isolates in hospitalized burn patients were gel E and asa genes in E. faecalis (48.5%) and E. faecium (43%). The cyl gene was not identified in any E. faecium isolates but in E. faecalis reported 5.8% [29]. In our study efa, A gene was the most frequent virulence (100%), ace (74%), gel E (70 %) and agg (52 %). Consistent with other reports, the results of the present study indicated that biofilm formation was higher in isolates with antibiotic resistance to Cotrimoxazole, Vancomycin, Ceftazidime and gentamicin suggesting a genetic association between the biofilm and these antibiotic genes. This conclusion was, however, not inclusive for all antibiotics [42, 43]. This finding shows that biofilm production in E. faecalis isolated from UTI in Kermanshah is higher than in other studies.
CONCLUSION
The high antibiotic resistance observed in this study likely suggests the inherent resistance of enterococci. The higher antibiotic resistance among the patient isolates could be attributed to the circulation of transposable elements carrying resistant genes in clinical isolates, indiscriminate and uncontrolled usage of antibiotics, and the presence of biofilm. In the present study, all isolates could produce biofilm. Therefore, a comparison was made between isolates producing strong, moderate, and weak biofilm, and no significant relationship was found between biofilm type and antibiotic resistance. Given the likely transmission of antibiotic resistance genes among enterococci and Staphylococci, managing the infections triggered by enterococci as well as the proper administration of antibiotics in patients can be good options to treat nosocomial infections effectively.
Abbreviations
Ace: Adhesin of collagen from E. faecalis
CLSI: Clinical and Laboratory Standards Institute
Esp: Extracellular surface protein
E. faecalis: Enterococcus faecalis
E. faecium: Enterococcus faecium
EBP: Endocarditis and Biofilm associated Pili)
Efa A: E. faecalis antigen A
UTI: Urinary Tract Infection
Declarations
Ethics approval and consent to participate
In the current study, we tried to protect the life, health, dignity, integrity, rights to self-determination, privacy, and confidentiality of personal information of research subjects. We conform to generally accepted scientific principles based on a thorough knowledge of the scientific literature, other relevant sources of information, and adequate laboratory and, as appropriate, animal experimentation. All samples were taken from volunteer patients for this research. All ethical issues were considered and this research was performed with hospitals’ permission. The name and identity, personal information and even patients’ illnesses and their medical information remained secret. All of the patients voice their satisfaction to use their sample in this investigation, especially to determine antibiotic resistance in UPEC strains
Consent to Publish
Not applicable.
Availability of data and materials
All data analyzed during this study are included in this published article.
Competing interests
The authors declare that they have no competing interests.
Funding
No funding was received.
Authors’ Contributions
MS, ET, HM, and RR carried out the molecular genetic studies, participated in the primers sequence alignment and drafted the manuscript. MS and HM carried out the sampling and culture method. MS and ET participated in the design of the study, performed the statistical analysis and writing the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
The authors would like to thank Manochehr Momeni Shahraki for his assistance in sample collection.
REFERENCES
- Bourgogne, Agathe, L. Charlene Thomson, and Barbara E. Murray. "Bicarbonate enhances expression of the endocarditis and biofilm associated pilus locus, ebpR-ebpABC, in Enterococcus faecalis." BMC microbiology, 2010; 10(1): 17.
- Yasuda, H., Y. Ajiki, J. Aoyama, and T. Yokota. "Interaction between human polymorphonuclear leucocytes and bacteria released from in-vitro bacterial biofilm models." Journal of medical microbiology, 1994; 41(5): 359-367.
- Kühn, Inger, Aina Iversen, Maria Finn, Christina Greko, Lars G. Burman, Anicet R. Blanch, Xavier Vilanova et al. "Occurrence and relatedness of vancomycin-resistant enterococci in animals, humans, and the environment in different European regions." Appl. Environ. Microbiol. 2005; 71(9): 5383-5390.
- Shahraki, Shahram, and Morteza Rabi Nezhad Mousavi. "Determination of virulence factors in clinical multidrug resistance Enterococci isolates at Southeast of Iran." Jundishapur Journal of Microbiology, 2017; 10(5).
- Nallapareddy, Sreedhar R., Kavindra V. Singh, Jouko Sillanpää, Danielle A. Garsin, Magnus Höök, Stanley L. Erlandsen, and Barbara E. Murray. "Endocarditis and biofilm-associated pili of Enterococcus faecalis." The Journal of clinical investigation, 2006; 116(10): 2799-2807.
- Nielsen, Hailyn V., Ana L. Flores-Mireles, Andrew L. Kau, Kimberly A. Kline, Jerome S. Pinkner, Fabrice Neiers, Staffan Normark, Birgitta Henriques-Normark, Michael G. Caparon, and Scott J. Hultgren. "Pilin and sortase residues critical for endocarditis-and biofilm-associated pilus biogenesis in Enterococcus faecalis." Journal of bacteriology, 2013; 195(19): 4484-4495.
- Haas, Wolfgang, Brett D. Shepard, and Michael S. Gilmore. "Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction." Nature, 2002; 415(6867): 84-8
- Elsner, H-A., I. Sobottka, D. Mack, R. Laufs, M. Claussen, and R. Wirth. "Virulence factors of Enterococcus faecalis and Enterococcus faecium blood culture isolates." European Journal of Clinical Microbiology and Infectious Diseases, 2000; 19(1): 39-42.
- Tille P. Bailey & Scott's Diagnostic Microbiology. Elsevier eBook on VitalSource.14th Edition. Elsevier Health Sciences; 2018.
- Winn, Washington C. Koneman's color atlas and textbook of diagnostic microbiology. Lippincott williams & wilkins, 2006.
- Gapeleh F, Mehrabi MR, Mirzaee M, Labibzadeh M. Biofilm formation and presence of Esp and cylA genes Enterococcus faecalis isolated from Hospital infection. Clin Microbiol and Case Reports. 2015; 1:1-4.
- Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing, CLSI document M100-S15,
- Sambrook J, Russell DW. Molecular cloning. Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor. 58-152.
- Hassan MM, El-Sayed BB. Antibiotic resistance and virulence genes in enterococcus strains isolated from different hospitals in Saudi Arabia. Biotechnology and Biotechnological Equipment. 2016; 30: 726-732.
- Barbosa, J., P. A. Gibbs, and P. Teixeira. "Virulence factors among enterococci isolated from traditional fermented meat products produced in the North of Portugal." Food Control, 2010(5): 651-656.
- Semedo, Teresa, Margarida Almeida Santos, Maria FátimaSilva Lopes, José J. Figueiredo Marques, Maria TeresaBarreto Crespo, and Rogério Tenreiro. "Virulence factors in food, clinical and reference enterococci: a common trait in the genus?." Systematic and Applied Microbiology, 2003; 26(1): 13-22.
- Medeiros, Aline Weber, Rebeca Inhoque Pereira, Daniele Vargas de Oliveira, Paula Dalcin Martins, Pedro Alves d'Azevedo, S. Van der Sand, Jeverson Frazzon, and Ana Paula Guedes Frazzon. "Molecular detection of virulence factors among food and clinical Enterococcus faecalis strains in South Brazil." Brazilian Journal of Microbiology 2014; 45(1): 327-332.
- Goudarzi, Mansour, Ashraf Mohabati Mobarez, Shahin Najar-Peerayeh, and Mohsen Mirzaee. "Prevalence of biofilm formation and vancomycin-resistant genes among Enterococcus faecium isolated from clinical and environmental specimens in Lorestan hospitals." Iranian journal of microbiology, 2018; 10(2): 74.
- Fisher, Katie, and Carol Phillips. "The ecology, epidemiology and virulence of Enterococcus." Microbiology, 2009; 155(6): 1749-1757.
- Sharifi, Yaeghob, Alka Hasani, Reza Ghotaslou, Behrouz Naghili, Mohammad Aghazadeh, Mortaza Milani, and Ahad Bazmany. "Virulence and antimicrobial resistance in enterococci isolated from urinary tract infections." Advanced pharmaceutical bulletin, 2013; 3(1): 197.
- Talebi, Malihe, Nastaran Asghari Moghadam, Zeynab Mamooii, Mohsen Enayati, Mahnaz Saifi, and Mohammad Reza Pourshafie. "Antibiotic resistance and biofilm formation of Enterococcus faecalis in patient and environmental samples." Jundishapur journal of microbiology, 2015; 8(10).
- Rashidan, Marjan, Zohreh Ghalavand, Gita Eslami, Latif Gachkar, Mohammad Rahbar, Ronak Khosravi, Ghazaleh Ghandchi, and Fatemeh Fallah. "Molecular detection of antibiotic resistance genes among Enterococcus faecalis isolated from fecal and urine samples of patients with community-acquired urinary tract infections." Archives of Pediatric Infectious Diseases, 2016; 4(3).
- Ramadhan, A. A., and E. Hegedus. "Biofilm formation and esp gene carriage in enterococci." Journal of clinical pathology, 2005; 58(7): 685-686.
- Tendolkar, Preeti M., Arto S. Baghdayan, Michael S. Gilmore, and Nathan Shankar. "Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis." Infection and immunity, 2004; 72(10): 6032-6039.
- Mozii F, Raya RR, Vignolo GM. Biotechnology of lactic acid bacteria: Novel applications. Int J Food Microbiol. 2010; 87: 532-555.
- Strompfová, Viola, Andrea Lauková, Monika Simonová, and Miroslava Marciňáková. "Occurrence of the structural enterocin A, P, B, L50B genes in enterococci of different origin." Veterinary Microbiology, 2008; 132(3-4): 293-301.
- Kafil, Hossein Samadi, and Ashraf Mohabati Mobarez. "Assessment of biofilm formation by enterococci isolates from urinary tract infections with different virulence profiles." Journal of King Saud University-Science, 2015; 27(4): 312-317.
- Gozalan, Aysegul, Fatma Filiz Coskun-Ari, Birsen Ozdem, Ozlem Unaldi, Nevreste Celikbilek, Fisun Kirca, Sibel Aydogan et al. "Molecular characterization of vancomycin-resistant Enterococcus faecium strains isolated from carriage and clinical samples in a tertiary hospital, Turkey." Journal of medical microbiology, 2015; 64(7): 759-766.
- Shokoohizadeh, Leili, Alireza Ekrami, Maryam Labibzadeh, Liaqat Ali, and Seyed Mohammad Alavi. "Antimicrobial resistance patterns and virulence factors of enterococci isolates in hospitalized burn patients." BMC research notes, 2018; 11(1): 1.
- Heidari, Hamid, Mohammad Emaneini, Hossein Dabiri, and Fereshteh Jabalameli. "Virulence factors, antimicrobial resistance pattern and molecular analysis of Enterococcal strains isolated from burn patients." Microbial pathogenesis, 2016; 90: 93-97.
- Sabia, Carla, S. De Niederhäusern, Elisa Guerrieri, Patrizia Messi, Immacolata Anacarso, Giuliano Manicardi, and Moreno Bondi. "Detection of bacteriocin production and virulence traits in vancomycin‐resistant enterococci of different sources." Journal of applied microbiology, 2008; 104(4): 970-979.
- Waar, Karola, Albrecht B. Muscholl-Silberhorn, Rob JL Willems, Maarten JH Slooff, Hermie JM Harmsen, and John E. Degener. "Genogrouping and incidence of virulence factors of Enterococcus faecalis in liver transplant patients differ from blood culture and fecal isolates." The Journal of infectious diseases, 2002; 185(8): 1121-1127.
- Heikens, Esther, Masja Leendertse, Lucas M. Wijnands, Miranda van Luit-Asbroek, Marc JM Bonten, Tom van der Poll, and Rob JL Willems. "Enterococcal surface protein Esp is not essential for cell adhesion and intestinal colonization of Enterococcus faecium in mice." BMC microbiology, 2009; 9(1): 19.
- Kristich, Christopher J., Yung-Hua Li, Dennis G. Cvitkovitch, and Gary M. Dunny. "Esp-independent biofilm formation by Enterococcus faecalis." Journal of bacteriology, 2004; 186(1): 154-163.
- Hällgren, Anita, Carina Claesson, Baharak Saeedi, Hans-Jürg Monstein, Håkan Hanberger, and Lennart E. Nilsson. "Molecular detection of aggregation substance, enterococcal surface protein, and cytolysin genes and in vitro adhesion to urinary catheters of Enterococcus faecalis and E. faecium of clinical origin." International Journal of Medical Microbiology, 2009; 299(5): 323-332.
- Dahlén, Gunnar, Susanne Blomqvist, Annica Almståhl, and Anette Carlén. "Virulence factors and antibiotic susceptibility in enterococci isolated from oral mucosal and deep infections." Journal of oral microbiology, 2012; 4(1): 10855.
- Seno, Yuko, Reiko Kariyama, Ritsuko Mitsuhata, Koichi Monden, and Hiromi Kumon. "Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract." Acta Medica Okayama 2005, 59(3): 79-87.
- Tendolkar, Preeti M., Arto S. Baghdayan, Michael S. Gilmore, and Nathan Shankar. "Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis." Infection and immunity, 2004; 72(10): 6032-6039.
- Toledo-Arana, Alejandro, Jaione Valle, Cristina Solano, Marı́a Jesús Arrizubieta, Carme Cucarella, Marta Lamata, Beatriz Amorena, José Leiva, José Rafael Penadés, and Iñigo Lasa. "The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation." Appl. Environ. Microbiol. 2001, 67(10): 4538-4545.
- Molinos, Antonio Cobo, Hikmate Abriouel, Nabil Ben Omar, Rosario Lucas López, and Antonio Galvez. "Detection of ebp (endocarditis-and biofilm-associated pilus) genes in enterococcal isolates from clinical and non-clinical origin." International journal of food microbiology, 2008; 126(1-2): 123-126.
- Zheng, Jin-xin, Bing Bai, Zhi-wei Lin, Zhang-ya Pu, Wei-ming Yao, Zhong Chen, Duo-yun Li, Xiang-bin Deng, Qi-wen Deng, and Zhi-jian Yu. "Characterization of biofilm formation by Enterococcus faecalis isolates derived from urinary tract infections in China." Journal of medical microbiology, 2018, 67(1): 60.
- Kayaoglu, Güven, and Dag Ørstavik. "Virulence factors of Enterococcus faecalis: relationship to endodontic disease." Critical Reviews in Oral Biology & Medicine, 2004, 15(5): 308-320.
- Park, Shin Yong, Kyoung Mi Kim, Joon Ha Lee, Sook Jae Seo, and In Hee Lee. "Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum." Infection and immunity, 2007, 75(4): 1861-1869.
